FAQ
No, the PMGC performs research services only.
The PMGC is interested in participating in collaborative research projects with academic and commercial groups. We are particularly interested in collaborations which can be funded by small or large collaborative grants. A key area of focus for us is in the area of protocol and technology development. If you are interested in a collaborative project with us, please contact Troy Ketela (click name to open email).
Our shipping address is:
Princess Margaret Genomics Centre
9-601, PMCRT
101 College St.
Toronto, ON, M5G 1L7
Samples should be shipped on dry ice. We highly
recommend shipping samples on a Monday or Tuesday to
provide enough time for the samples to arrive before the
weekend and helping to assure the dry ice will not
sublime before arrival. Please note that special
paperwork is required for shipping RNA or DNA from
locations outside of Canada. For the Customs
documentation that you will need, please download this
form
. Once you send us this information, the UHN Customs
Officer will generate the appropriate Customs document
and send it to you via e-mail. This document must be
included with the waybill on your package.
| Bioanalyzer kit | Amount requested | Concentration range |
|---|---|---|
| RNA Nano | 2-3 µL total RNA in RNase-free water | 100-200 ng/µL |
| RNA Pico | 2-3 µL total RNA in RNase-free water | 5-10 ng/µL |
If you do not know the concentration of your sample, you will be charged for running the sample even if the concentration is too low to detect. We kindly ask that you dilute concentrated samples accordingly as high concentrations can cause problems with the assay.
The Agilent Bioanalyzer can also be used to measure mRNA, amplified RNA, labelled-cDNA, and other nucleic acids, however, the standard service we are offering currently applies to total RNA only. If you have specific needs, please contact us .
Assessing the quality of your RNA sample before proceeding with labelling, etc, can save a lot of time and money. It can also reduce the time spent troubleshooting if something goes wrong at the next step. If you know your RNA was good, you can rule out degraded RNA as the cause of your problem.
The Agilent Bioanalyzer is a convenient replacement for standard denaturing agarose gels; it is much faster, requires only a small amount of sample, and is more environmentally-friendly. The Bioanalyzer uses capillary electrophoresis (lab-on-a-chip technology) to move the sample through a gel matrix. An intercalating dye in the matrix allows the nucleic acid to be detected as it moves through the capillary. The electropherogram provides the user with a detailed trace of each sample.
The RNA Integrity Number (RIN) is a software tool that enables users to estimate the integrity of total RNA samples. The RIN is a numerical assessment of the integrity of RNA and allows for the direct comparison of RNA samples.
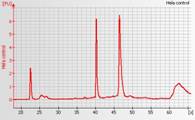
High quality total RNA
The electropherogram should have 2 distinct peaks around 41 and 47 seconds (corresponding to the 18S and 28S RNA), as well as the marker around 22 seconds, and the baseline should be flat.
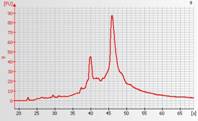
Partially degraded total RNA
The presence of a broad peak between 23-30 seconds indicates the presence of small (degraded) RNA products and tRNA. When RNA is partially degraded, the 18S and 28S peaks are often not very distinct and the baseline is raised.
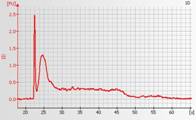
Degraded total RNA
Total RNA is completely degraded when distinct 18S and 28S peaks are not visible on the electropherogram.
For nucleic acids, the minimum volume required is 1 µl and the concentration should be between 2-3700 ng/µl (NanoDrop) and either 0.1-1000 ng/µl for dsDNA or 0.25-100 ng/µl for RNA (Qbit). For protein samples, 2 µl of sample is required.
Customers are asked to bring/submit at least 2 µl for nucleic acid quantitation (NanoDrop or Qbit) and 4 µl for protein quantitation (NanoDrop only).
No, the sample cannot be retrieved following its measurement.
The NanoDrop ND-1000 spectrophotometer is able to perform full spectrum UV-Vis absorbance analyses (220-750 nm). Using the microarray application, the concentration of nucleic acid and up to 2 dyes can be calculated and displayed at the same time.
There are a few solvent restrictions when using the NanoDrop. For example, hydrofluoric acid should definitely not be used. Samples dissolved in high vapour pressure solvents such as acetone can be measured but only using the short path length to reduce evaporation. Please contact us prior to using the NanoDrop if you are using non-standard buffers/solvents.
Deep sequencing requires that every base in a sample is sequenced several times for two reasons. First, it is necessary to gather multiple observations per base to generate a reliable base call and secondly, reads are not distributed evenly over an entire genome, due to the random nature of the sequence generation - some bases will be read many more times than the average but some will be read far fewer times. 'Depth of sequencing' refers to the number of times a genome has been sequenced.
There is not a definite answer to this question. Most
users choose the coverage level required for a
particular experiment by considering the following
factors: the type of study, gene expression level, size
of reference genome and published literature.
For example, for studies of human genome
mutations/SNPs/rearrangements coverage of 10-30 times
depth of coverage depending on the application and
statistical model. Whereas ChIP-seq studies, where only
a subset of a genome is being investigated, generally
require around 100 times coverage.
For RNA-seq determining the level of coverage required
is further complicated by the varied expression levels
of different transcripts. Highly expressed transcripts
will be read many more times than transcripts with low
expression. For RNA-seq experiments, it is usually the
number of millions of reads required that is considered.
The number of reads required will depend on how
sensitive the experiment needs to be for genes of
low-level expression.
Illumina provides a
Coverage Calculator
that might aid you in determining the level of coverage
your experiment requires.
The
ENCODE Consortium
also provides some useful
data standards
, including these
guidelines
for experiments which address many important
considerations for deep sequencing experiments.
With ChIP-seq generally there is a lower requirement for
number of reads than in the case of RNA-seq and this
makes ChIP-seq as attainable as a ChIP-chip experiment.
According to the ENCODE ChIP-Seq standards the current
minimum standard number of reads for a ChIP-seq
experiment, involving transcription factors and
chromatin modifications, is 10 million uniquely mapped
reads per replicate for mammalian cells.
As with any technique, replicates are necessary for statistical assessment of significant differences between groups. However, due to the quantitative nature of sequencing, useful information can also be obtained from singular experiments, in such instances as case studies where conclusions are being applied only to the samples being tested and not to any groups. Consultation will be required for determining the required number of replicates for a particular set of experiments.
The PMGC uses a series of processing steps, or
algorithms, that are based on current publications in
the field, for processing all sequencing data. This
series of steps is referred to as the "pipeline". The
pipeline will vary depending on the type of analysis
required (RNA-seq vs. Exome-seq vs. ChIP-seq etc.). For
more details please inquire.
Customers will receive their processed sequence data as
.fastq files along with FASTQC data. If alignment or
further analysis is performed by the PMGC then the form
the results take will depend on the type of analysis
required. Please inquire.
We offer a number of different platforms to help meet most applications. For example for researchers wishing to study highly heterogeneous samples, the 10x Genomics Chromium system allows for the analysis of 1,000 to 10,000 cells per sample, and requires at least 50,000 cells to be provided to us (although, we can work with smaller numbers of cells in specific situations). Alternatively, for highly homogeneous cell populations, the Fluidigm C1 or our Direct Sort applications can analyze up to 96 cells with only 2,000 cells as the initial input.
The answer is dependent on the technology platform being used. The 10x Genomics approaches analyze short (~90 base) reads located near the 3’ or 5’ ends of transcripts. This leads to a more cost-effective analysis of gene expression from thousands of cells. For researchers wishing to look at the full transcripts, the Fluidigm C1 system or plate-based approaches use Clontech's SMARTer V4 chemistry to analyze full length transcripts.
The average number of genes detected per cell really depends on the cell type. Large, fast growing cells like those from culture or from tumours will often have 5,000 or more genes detected per cell on average. Primary immune cells that are smaller, and less transcriptionally active may only have ~900 to 3,000 genes detected per cell.
Global RNA amplification protocols do have some inherent sensitivity limitations which does cause transcripts of low abundance to be missed on occasion from single cells. The PMGC also offers single cell qPCR methods with the Fluidgim C1 coupled to the Fluidgim BioMark HD. In this application 96-192 genes can be detected with high sensitivity for up to 96 individual cells.
Unfortunately, due to the specialized nature of the instruments as well as the importance of them to our operation, we do not allow researchers outside our core to operate the instruments themselves. If you would like to see the instruments to better understand how the techniques work, we'd be happy to have you come by for a visit.
Absolutely - we offer free consultation to any potential customer. We are here to help! Please contact us and we'd be happy to meet in person, by phone or by virtually.
Freshly mounted sections are preferred for Visium. Please consult with our team.
10X now supports immunostaining with fluorescently labeled antibodies for Visium workflows.
Because selection of the precise sections that you’ll want to use for Visium depends on a number of factors, we encourage clients to discuss their plans with us well in advance. In general, we’ll suggest that clients select and mount their own sections on Visium slides that either they have purchased or that we provide. It is possible for us to mount sections that you have made that are not yet mounted.
Yes, sections mounted on Visium slides are stable for up to 4 weeks, during which you can ship them on dry ice to us for processing. Please consult with us prior to mounting your sections for advice and shipping recommendations.
Currently, we do not offer DNA, RNA or protein extraction services for samples destined for Nanostring or Luminex platforms.
There are three main differences between these two products:
Difference in Build: A Custom CodeSet has the gene-specific sequences built in to the reporter and capture probes whereas a TagSet would require the user to order gene-specific oligonucleotides.
Level of Multiplexing: A Custom CodeSet can multiplex up to 800 genes and a TagSet up to 192 targets.
Difference in Workflow: If ordering a Custom CodeSet, NanoString will provide all of the probes necessary for the reaction at the appropriate concentrations. For a Custom TagSet, NanoString provides the recommended sequence information, and you will need to order the oligonucleotides and prepare them at recommended concentrations before adding them to the hybridization reaction.
The earlier you consult with us in your experimental design, the better we can help meet your research needs. We will work with you to design the strategy for your work plan – from custom panels and catalog assay selection, how to best store your samples for long term. We will provide quotes upon which you can base your funding requests and operating budget.
Luminex licenses the rights to create xMAP ® assays to a number of xMAP ® partners, who in turn create a number of different assays with varying content. As the nature of these assays is to focus on specific subsets of proteins or genes, each assay provider has developed different content, which taken in total gives a fairly broad spectrum of potential analytes to profile.
Since Luminex licences the technology, they are able to keep track of all of the assays that are generated. Luminex has created an excellent tool called xMAP ® Kit Finder which can be used to determine if any assays exist for your analyte of interest. Note that if you use this tool, choose the Luminex 100/200 instrument option if you would like PMGC to provide the service for you.
The amount of sample required is dependent on the type of assay being performed. The numbers provided here are guidelines only. For more accurate information on sample requirements please contact us.
For Cytokine assays:
- Plasma: 15 µl
- Serum: 15 µl
- Cell culture media: 55 µl
For Phosphoprotein assays:
- Cell culture or tissue samples should be prepared with the appropriate cell lysis kit (such as the Bio-Rad Cell Lysis kit for Bio-Plex assays).
- 55 µl of protein lysate in assay buffer should be provided at a concentration of about 400 µg/µl. Please contact us for further guidance.
Assays based on the xMAP ® technology use a liquid suspension array with up to 100 uniquely colour-coded bead sets. Each of the 100 bead types are internally labelled with a specific ratio of two fluorophores to assign it a unique spectral address. The beads are then conjugated with different biomolecules (including RNA, DNA, enzyme substrates, receptors, antigens, and antibodies), allowing the capture of specific analytes from the sample. A fluorescently-labelled reporter molecule is then added to the sample in order to detect and quantitate each captured analyte. The beads are drawn through a flow cell where two lasers excite each bead. Fluorescent signals are recorded, translating the signals into data for each bead-based assay. To see a video description of how the Luminex xMAP ® technology works please click here .
Yes! The Bio-Plex system at the PMGC can also run xMAP assays available from any xMAP ® certified vendor including:
Some Luminex assays are designed to be used in quantitative manner. For example, most of the cytokine assays, provided by vendors such as Bio-Rad and Millipore, are packaged with a set of standards that can be used to generate a standard curve allowing for quantitation of the individual analytes in a sample. Usually we generate an 8 point standard curve that will have a dynamic range of 3 to 5 logs depending on the individual analyte. Please note, in order to make the assay quantitative, 16 measurements must be made of these standard controls, leaving 80 assays available per plate for your experimental samples.
Other assays such as the phospho-protein assays tend to be relative – whereby the degree of phosphorylation is compared between samples in a ratiometric manner rather than a fully quantitative manner.
A biological replicate involves independent samples (multiple patients, multiple biopsies from an individual patient, etc). Serum or plasma from multiple patients would be profiled independently, or cell culture media from multiple individual cell culture dishes would be assayed. The purpose of a biological replicate is to assess and control for biological diversity.
A technical replicate involves splitting a sample at some point and continuing on with the two aliquots through the rest of the protocol. So for example, a technical replicate might involve taking one serum sample and performing two independent assays from that initial sample. Technical replicates provide an indication of measurement (or technical) error, and are useful for diagnosing problems with the protocol but offer little in the way of statistical power for a biological experiment.
The exact number of replicates required for an experiment is difficult to determine a priori without a proper power analysis. Such a power analysis is not always possible as it requires that you have an estimation of the overall variance, which you often do not have before you perform the experiment. We generally recommend doing as many biological replicates as your budget can accommodate. In general, it is good to have at least three biological replicates per condition. For a more detailed determination of the number of replicates required please contact us as we will be happy to help you design your experiment.
The fact that many beads are read per sample, in a single assay is an example of technical replication. This level of “oversampling” ensures that that one particular measurement of a specific analyte in a specific sample is relatively accurate. However, this does nothing to address the biological diversity of a particular response.
While each sample is measured relatively accurately, multiplex protein assays are complex and are much more chemically diverse than a nucleic acid assay such as a microarray. Due to the complexity and occurrence of phenomena such as “matrix effects”, it is often a good idea to measure each sample in duplicate or triplicate (as a technical replicate). Of course, this is dependent on both your budget and the amount of sample you have available.
It is more important to provide a sufficient number of biological replicates (i.e. serum from a large number of patients) in order to provide sufficient statistical power for your experiment.
As with any multiplexed assay, there is a level of data analysis that must be performed to help filter noise, perform statistical comparisons, and ultimately to determine the biological meaning of the data. The Bio-Plex Manager v5.0 software provides system control, validation, calibration, data acquisition, and data analysis for multiplex assays. The software will calculate concentration, standard deviation, coefficient of variation, and percent recovery. A standard data normalization is carried out using internal controls to allow for comparison between plates, experiments, and samples. Further data analysis is possible including higher level statistical analysis.
Multiplexed assays such as Luminex assays are relatively complex, with numerous steps, reagents and instruments involved. Despite this, it has been shown in many studies that the leading cause of variance is the facility that is performing the analysis, followed by the technician that completes the experiment. All of our technicians are highly experienced, and rigorously trained. Each project is assigned a specific technician, who completes the entire project to minimize variability. Furthermore, wherever possible, we use assays and reagents from the same lot. We use the same equipment (plate washer, incubator, plate shaker) throughout the project, all in an effort to ensure the tightest possible data.
Often a project is large enough that it will require multiple days worth of labwork to complete. In these cases we work with the customer to identify which samples are replicates and we split these replicates across the various days to ensure that it is possible to control for any day-to-day variations that may occur.
An “analyte” is a specific protein biomarker of interest which you can quantify using Luminex assay kits that are commercially available.
Each kit is developed and validated to perform specifically with the reagents included and at the suggested sample dilutions. Combining kits can risk unwanted antibody cross-reactivity, incompatible serum matrices and skewed results.
PMGC uses The Luminex ® 200 ™ instrument and it uses a flow cytometry-based system where beads flow in sheath fluid through a cell with a red laser (635 nm) identifying the bead region and a green laser (525 nm) quantifying the bound analyte “sandwich.” The MAGPIX ® instrument uses a CCD imager to identify beads and quantify analytes with red LED (635 nm) and green LED (525 nm) light sources.
The earlier you consult with us in your experimental design, the better we can help meet your research needs. We will work with you to design the strategy for your work plan – from biomarker and assay selection, how to best store your samples for long term. We will provide quotes upon which you can base your funding requests and operating budget.
For most of our applications, we'll require 300,000 cells per sample. This however is flexible and if your input amount doesn't quite reach this, please don't hesitate to contact us.
We can work with extracted DNA, fresh single cell suspensions, viably frozen single cell suspensions or frozen blocks of tissue.
For fresh samples, we ask customers to bring us the cells in a single cell suspension, in the media they're most happy in.
For frozen samples, we ask customers to bring us a block of tissue (minimum 30mg) in dry ice. For those who can't provide 30mg, please reach out to us.
Typical turnaround times for sequenced data these days range from 3-5 weeks due to pandemic scheduling.
It's okay! Just talk to us and we can try to figure it out together :D
How much money do you have? Typically 2 replicates are a minimum, with 3 being the normal amount of replicates to have.